About Genes Like Me (formerly GeneDecks Partner Hunter)
Genes Like Me is a novel analysis tool which provides a similarity metric by highlighting shared descriptors between genes, based on the rich annotation within the GeneCards compendium of human genes.
Genes Like Me Algorithm
Genes Like Me calculates similarity scores between each query gene and all remaining candidate genes in the GeneCards database for 8 attributes that appear in table 1. For all attributes except Gene Ontology, and sequence paralogy, the similarity score between a query gene and a candidate gene is calculated in the following manner: each descriptor score (DS) is the result of dividing its rank by Log10 of its frequency in the database
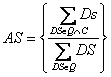 For the sequence paralogy attribute, if a partner candidate is also identified as a sequence paralog (SP),
then it is assigned a value of 1 for this attribute and 0 otherwise
For the sequence paralogy attribute, if a partner candidate is also identified as a sequence paralog (SP),
then it is assigned a value of 1 for this attribute and 0 otherwise
The attribute score is then multiplied by the weight given for the attribute and all attribute scores are then summed to give the Genes Like Me score (PHS)
Table 1
The attributes used in Genes Like Me algorithms with their contributing data sources.
| Attribute | Data Source |
|---|---|
| Sequence paralogy |
|
| Domains |
|
| Super Pathways |
|
| Expression patterns |
|
| Phenotypes |
|
| Compounds |
|
| Disorders |
|
| Gene Ontology |
|